npj Digital Medicine (CAS Q1 Top journal, 5-year Impact Factor ≈16.6) recently published the latest research titled “Large-vocabulary segmentation for medical images with text prompts,” conducted by the Smart Healthcare Team of the SJTU School of Artificial Intelligence.
In this work, the team introduced SAT (Segment Anything in Radiology Scans, driven by Text prompts) — the first 3D medical image segmentation foundation model that incorporates multimodal anatomical knowledge and is driven by text prompts. Its effectiveness and generalization capability were validated across a large-scale benchmark comprising 72 datasets.
This work (i.e., SAT) has been selected as an official baseline for the CVPR 2025 BiomedSegFM Challenge.
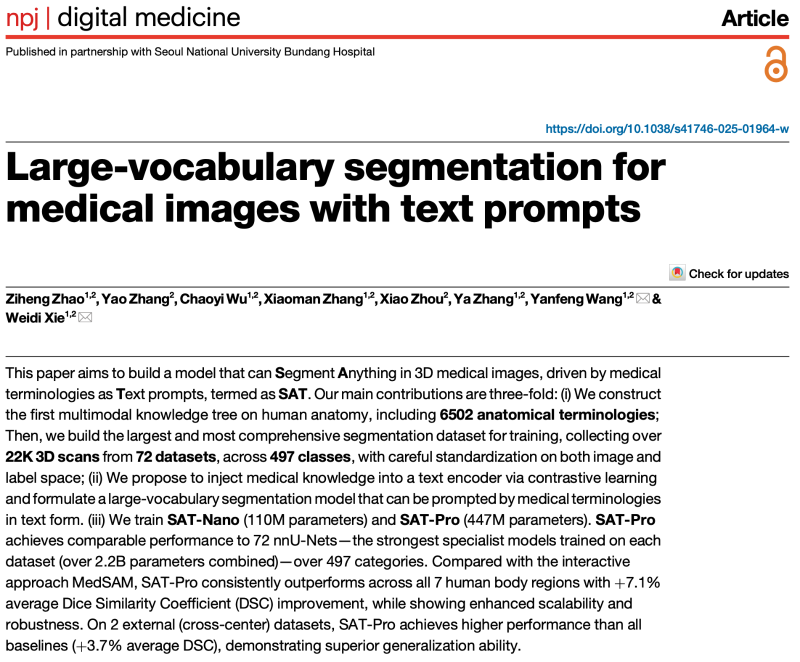
Medical image segmentation is vital for diagnosis, surgical planning, and disease monitoring. Traditional approaches rely on “specialized models” trained separately for each task, lacking generalizability across organs and imaging modalities.
Meanwhile, as large language models evolve rapidly in healthcare, there is an urgent need for segmentation tools that integrate language understanding with spatial localization.
To address these challenges, the SJTU team proposed SAT(Segment Anything in radiology scans, driven by Text prompts) — the first 3D segmentation foundation model enhanced by anatomical knowledge and guided by medical text prompts.
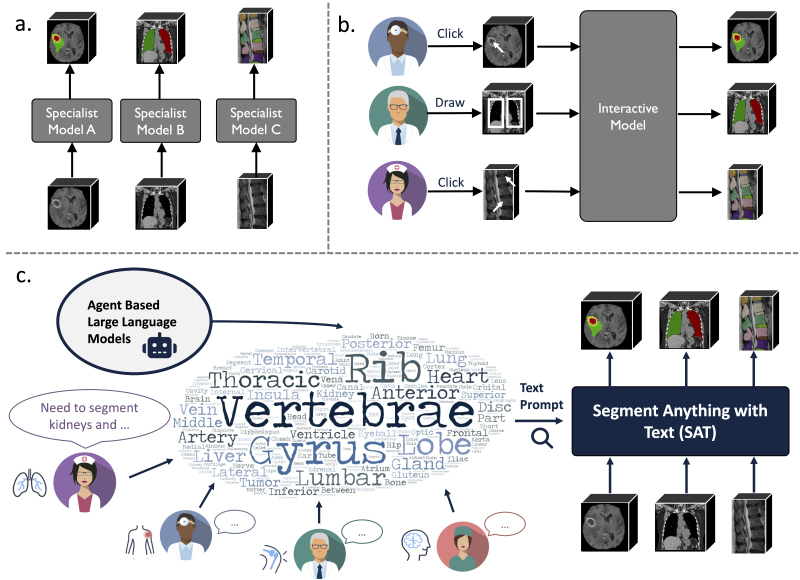
Figure 1: Differences in paradigm between SAT and existing specialized models (e.g., nnU-Net) and interactive models (e.g., MedSAM).
In terms of data, the research team first constructed the field’s first multimodal knowledge graph covering more than 6,000 human anatomical concepts, drawing on UMLS and several other sources. The graph includes detailed textual descriptions of each concept, multiple types of semantic relationships among concepts, as well as selected visual examples and segmentation annotations.
The team also integrated 72 public datasets to build SAT-DS, a large-scale 3D medical image segmentation dataset containing 22,186 3D scans and 302,033 segmentation annotations. SAT-DS spans three imaging modalities—CT, MRI, and PET—and covers 497 segmentation targets across eight major anatomical regions.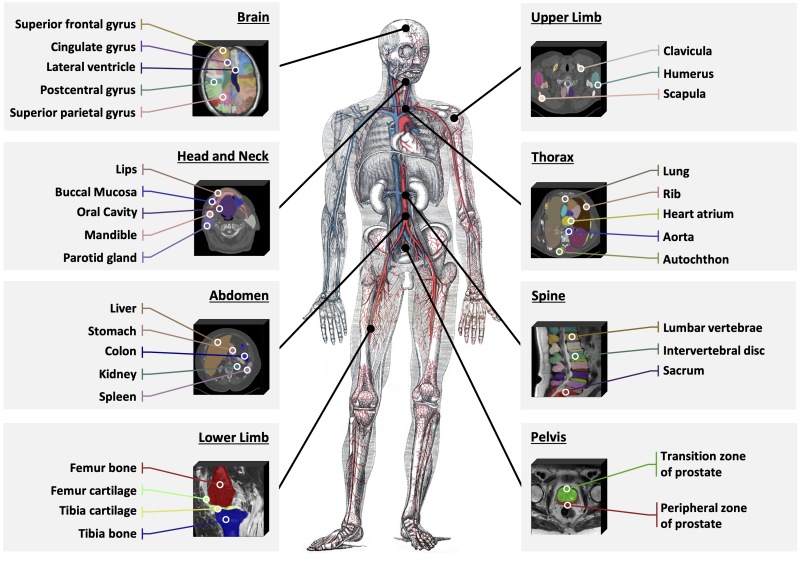
Figure 2: As one of the largest 3D medical image segmentation datasets in the field, SAT-DS covers 497 segmentation categories across eight major anatomical regions.
Methodologically, the research team proposed a text-prompt-driven segmentation framework that adopts a two-stage process of “knowledge injection+segmentation training.” First, multimodal anatomical knowledge from more than 6,000 concepts is injected into the text encoder through contrastive learning to obtain more accurate and visually aligned concept representations. Then, the encoded text prompts are combined with 3D image features to decode and produce segmentation predictions.
Figure 3: The construction of SAT consists of two stages: knowledge injection and segmentation training.
The research team trained two variants of the model on SAT-DS: SAT-Pro (447M parameters) and SAT-Nano (110M parameters). Across all 497 categories, SAT-Pro achieves performance comparable to 72 individually trained nnU-Net models, while reducing the parameter count by more than 80%, significantly lowering usage and deployment costs.
Compared with the interactive segmentation model MedSAM, SAT-Pro demonstrates substantial advantages in organ segmentation and can achieve fully automatic segmentation through text prompts.
Relative to contemporary work, BiomedParse (first released in May 2024), SAT-Pro delivers notably superior overall performance and supports a much larger vocabulary of segmentation categories.
The team also conducted external validation experiments, confirming SAT’s strong generalization ability across different datasets. More experimental details are provided in the published paper.
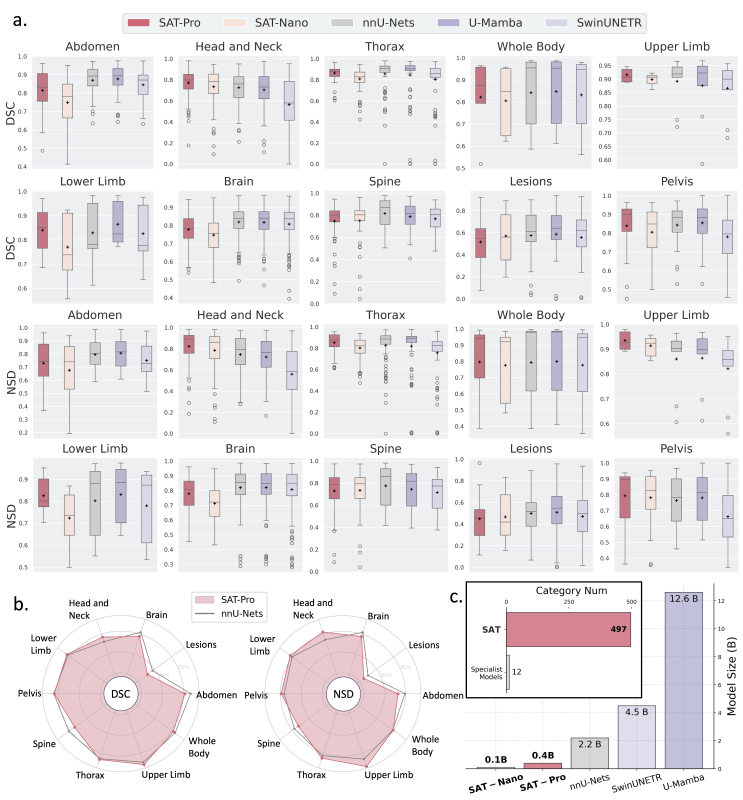
Figure 4: In-domain comparison of SAT-Pro and SAT-Nano with specialized models including nnU-Net, U-Mamba, and SwinUNETR.
As the first text-prompt-based 3D medical image segmentation foundation model, SAT offers value in multiple dimensions:
- A flexible and efficient foundation model: SAT achieves performance comparable to specialized models across diverse tasks using a single unified model, while drastically reducing parameter size and deployment cost.
- Fully automatic segmentation via text prompts: SAT eliminates the need for manual interaction required by models such as MedSAM.
- Scaling laws remain effective: Experiments show that increasing model size significantly improves performance on both internal and external validation sets.
- Domain knowledge enhancement improves performance: Incorporating anatomical knowledge notably improves segmentation accuracy, especially for long-tail categories.
- A natural agent for large language models: SAT can serve as a tool for LLMs, seamlessly integrating segmentation and localization capabilities through text, advancing the development of general-purpose medical AI.
This study was conducted by the Smart Healthcare Team of the SJTU School of Artificial Intelligence. The first author is Ziheng Zhao, PhD student at Shanghai Jiao Tong University. The corresponding authors are Prof. Yanfeng Wang and Assoc. Prof. Weidi Xie of the SJTU School of Artificial Intelligence. The work was supported by the Science and Technology Innovation 2030 “New Generation Artificial Intelligence (2030)” Major Project (2022ZD0160702).
All code, models, and datasets from this study have been open-sourced:
Code: https://github.com/zhaoziheng/SAT/
Paper: https://www.nature.com/articles/s41746-025-01964-w
Dataset: https://github.com/zhaoziheng/SAT-DS/
Author: SJTU School of Artificial Intelligence
Contributing Unit: SJTU School of Artificial Intelligence
Translated by: Denise
Proof read: Rui Su


