Assoc. Prof. Jun Ni’s research team from SJTU School of Life Sciences and Biotechnology/Synthetic Science Innovation Research Center of China Zhangjiang Institute for Advanced Study published a paper in Cell on January 24, 2025, titled Rational Multienzyme Architecture Design with iMARS, revealing the “code” of multienzyme spatial proximity and efficiency. This work solves the problem of no standard for multienzyme assembly for more than 50 years, and realizing the rational design of artificial multienzyme complexes from sequence to function.
Biocatalysis plays a crucial role in cellular metabolism and biomanufacturing, and multi-enzyme cascade catalysis is widely used in medicine, food and industry. In nature, the spatial structure of multienzyme complexes is often finely tuned to control the catalytic reaction rate. Inspired by that, researchers have constructed artificial multienzyme complexes by fusing enzymes, protein scaffolds, and nucleic acid scaffolds, among other assembly strategies, to improve the catalytic efficiency of cascade reactions. However, the spatial structure-activity relationships of artificial multienzyme complexes have plagued the academic community for more than 50 years, and the assembly of artificial multienzymes can only rely on experimental trial-and-error, and there is no corresponding rational design tool yet, which limits its application.
In order to address the above challenges, the research team analyzed the spatial structure-activity relationships of artificial multienzyme complexes through high-throughput testing and spatial structure prediction, and preliminarily deciphered the spatial proximity and efficiency of multienzymes, and found that the spatial distance of the fusion enzyme and the angle of the channel are the key factors affecting the catalytic efficiency. Based on this, the research team developed iMARS, a rational design tool for artificial multienzyme complexes, with a built-in database containing thousands of linkers of different properties, high-throughput protein structure was predicted through ParaFold, and calculated based on molecular docking and tools like CAVER, etc. and scored and screened with DO Score, which help to realize a rapid prediction of the relative activities from the amino acid sequences to the relative activities of the artificial multienzyme complexes.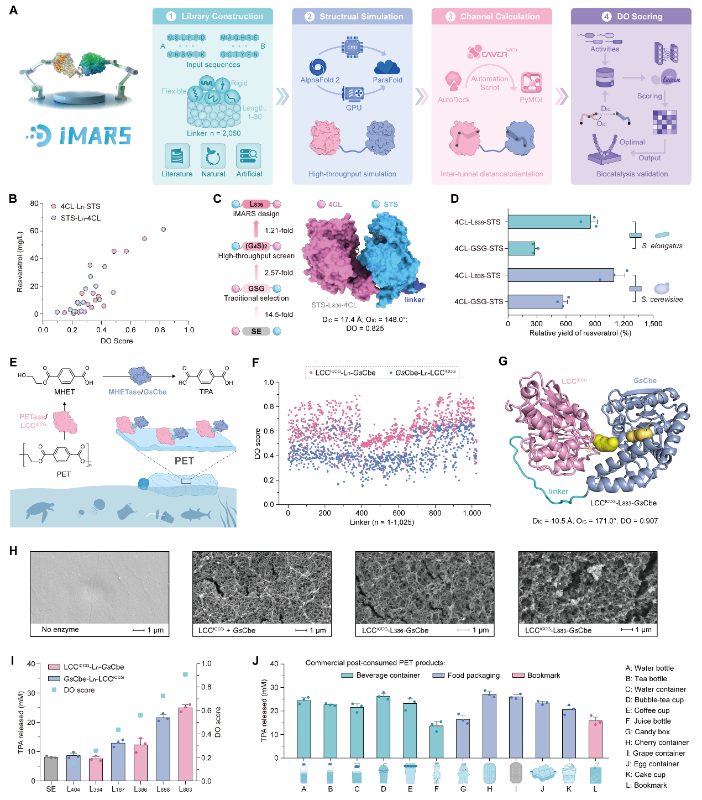
The team explored the design capabilities of the iMARS approach in different application scenarios in the biosynthesis of compounds such as resveratrol, vanillin, and ergothioneine, as well as in the biodegradation of PET plastics. In particular, compared to the free enzyme, the optimal fusion enzyme designed resulted in a 45.1-fold increase in production of resveratrol, an engineering E. coli, and was also applied in saccharomyces cerevisiae and photosynthetic microorganism cyanobacteria, demonstrating that iMARS as a molecular-level design tool can be widely applied to different host cells. In the in vitro biodegradation experiments of PET plastic, scanning electron microscopy (SEM) observed that the designed fusion enzyme degraded the PET film with more significant erosion marks. During the fed-batch fermentation for ergothioneine synthesis, the engineered strain containing the optimized designed fusion enzyme achieved the highest level of productivity, demonstrating the potential of iMARS in the industrialization of biomanufacturing. Currently, iMARS is available online (http://www.imars-lumybio.com/), facilitating researchers to rapidly design multienzyme complexes.
This research is based on a variety of deep learning software development, reflecting the role of artificial intelligence in advancing biology. The process of “creating, learning and designing” multienzyme complexes also reflects the concept of synthetic biology, which is “Creating to accumulate knowledge, creating for use”. The iMARS tool developed in this research is the first rational design tool for artificial multienzyme complexes, marking a new era of rational design from experimental trial-and-error method, which will promote the rapid development of synthetic biology and biomanufacturing, and is of great significance to the fields of medicine, food and industry.
Associate Professor Jun Ni from the School of Life Science and Biotechnology, State Key Laboratory of Microbial Metabolism, and Synthetic Science Innovation Research Center of China Zhangjiang Institute for Advanced Study, Shanghai Jiao Tong University (SJTU) is the corresponding author of the paper. Jiawei Wang, a doctoral student at SJTU, is the first author of the paper, with SJTU as the first institutional affiliation. Professor Yilei Zhao of SJTU and Shiyu Meng of Shanghai Lumybio Technology Co. Ltd. provided important support for this research. Additionally, this research was supported by the National Natural Science Foundation of China, the National Key Research and Development Program of the Ministry of Science and Technology, and Shanghai Lumybio.
Paper link: